- 10 Posts
- 3 Comments
Joined 1 year ago
Cake day: June 22nd, 2023
You are not logged in. If you use a Fediverse account that is able to follow users, you can follow this user.

 2·1 year ago
2·1 year agoDamn, that’s cool.
Here’s the same 3 morphogen periodic clock network implemented in python https://diode.zone/w/9ApBpVcU5CnCbbTu3Lcegw


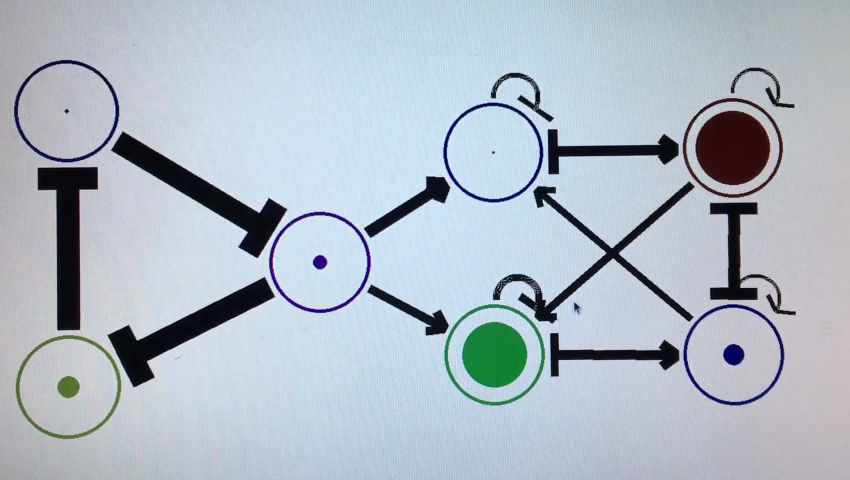
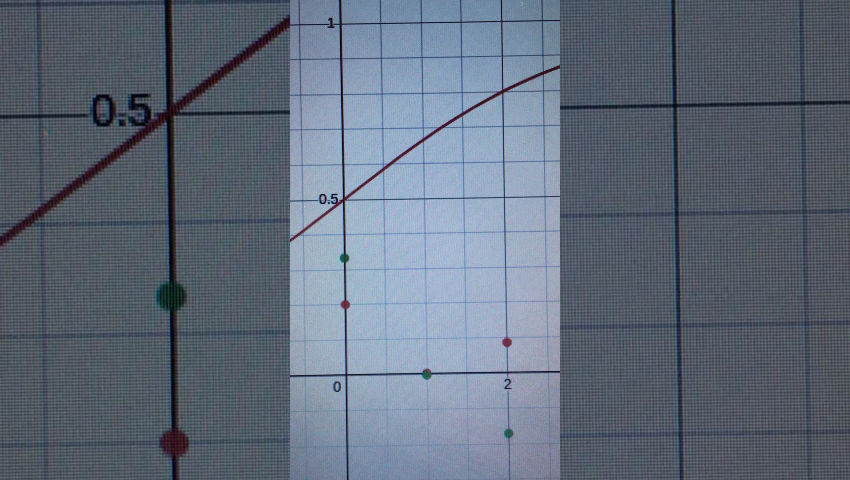



Well, it is technically differential equations, but with weighted inputs like a NN. Here’s the equations
for each node (gene/morphogen) i. zi is the concentration of morphogen i, zj is that of j. f(x) is the sigmoid function, k1 is the maximum rate of expression, k2 is the degradation rate, b is the bias. wij is the weight for an edge from j to i.
This is just written in python, so the network is defined by a matrix with each number representing the weight between two of the edges. I ignore the edge if it’s weight is zero.
What are the standard symbols for genetic circuits?
edit: sorry it’s impossible to see the equations if you have a black background.