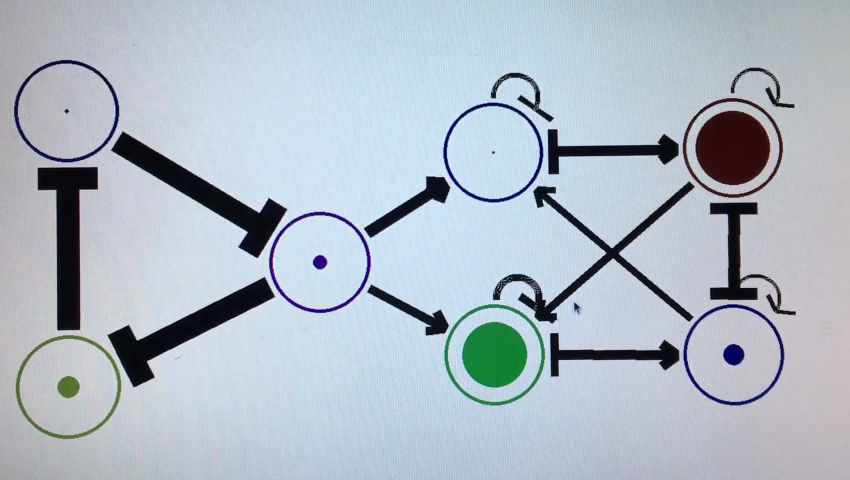
I’m working on a program to edit and simulate gene regulatory networks using recurrent neural networks as a model. Here’s a demo.
This is pretty cool, but is there a reason to use NN rather than differential equations? Seems like it might be more computationally expensive.
Are you using some kind of declarative language to define the networks? Not sure if a standard for that exists yet. There are the standard symbols for genetic circuits though.
Well, it is technically differential equations, but with weighted inputs like a NN. Here’s the equations


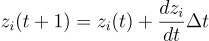
for each node (gene/morphogen) i. zi is the concentration of morphogen i, zj is that of j. f(x) is the sigmoid function, k1 is the maximum rate of expression, k2 is the degradation rate, b is the bias. wij is the weight for an edge from j to i.
This is just written in python, so the network is defined by a matrix with each number representing the weight between two of the edges. I ignore the edge if it’s weight is zero.
What are the standard symbols for genetic circuits?
edit: sorry it’s impossible to see the equations if you have a black background.